Introduction
KNN imputation is a widely used technique for handling missing data.
The idea is simple: for each missing value, find the k nearest neighbors based on available features and impute using their values.
While scikit-learn provides a convenient KNNImputer, it becomes prohibitively slow on large datasets due to its single-threaded implementation.
When working on ehrapy, our framework for electronic health record analysis, we encountered exactly this bottleneck - imputing 50,000 patient records with only 10% missing data took over 4 minutes.
This motivated me to build fknni, a drop-in replacement that achieves up to 500x speedup by leveraging FAISS and cuML.
Results
We benchmarked four implementations: scikit-learn's KNNImputer, our FastKNNImputer with FAISS (CPU and GPU), and the cupy/cuML backend.
All experiments used k=5 neighbors, 10% missing values, and the weighted imputation strategy.
GPU benchmarks ran on an NVIDIA A100.
Accuracy
All implementations produce comparable results, with our FAISS-based approach achieving slightly lower mean absolute error against the original data:
| Implementation | MAE vs Original |
|---|---|
| sklearn | 53.57 |
| FAISS CPU | 52.74 |
| FAISS GPU | 52.74 |
| cupy/cuML | 52.78 |
The small differences between sklearn and our implementations stem from distance computation precision and tie-breaking behavior. FAISS CPU and GPU produce identical results, while cuML shows minor variation due to float32 internal computations.
Performance
The performance gains are dramatic, especially at scale:
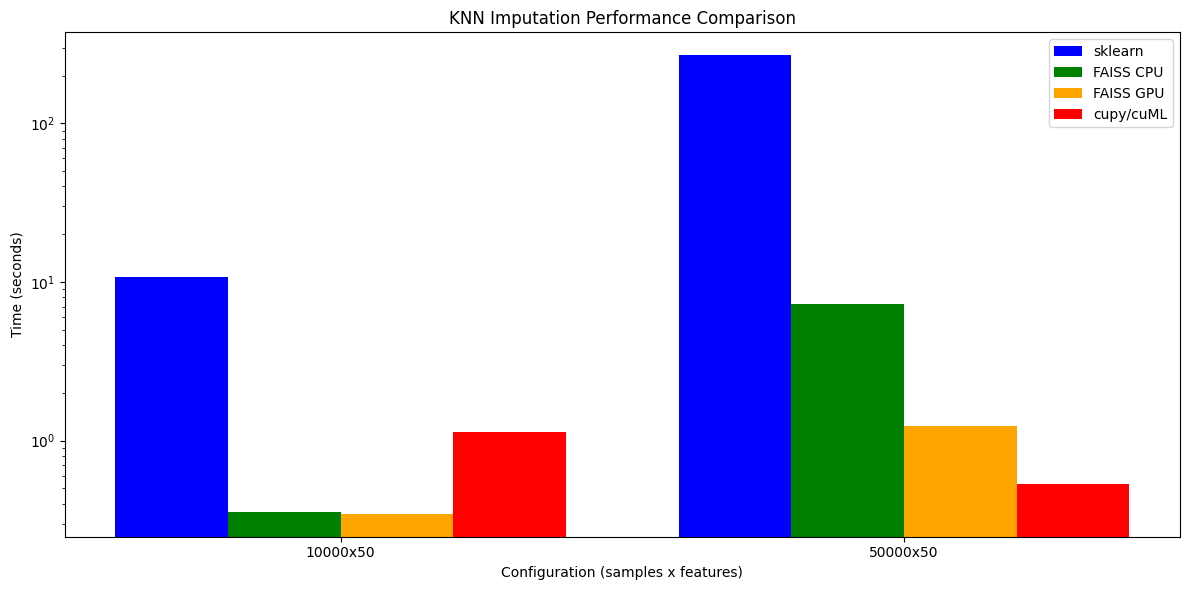 Speedup over sklearn KNNImputer across different configurations. At 50k samples, cupy/cuML achieves 507x speedup on A100.
Speedup over sklearn KNNImputer across different configurations. At 50k samples, cupy/cuML achieves 507x speedup on A100.
| Configuration | sklearn | FAISS CPU | FAISS GPU | cupy/cuML |
|---|---|---|---|---|
| 10k × 50 | 10.7s | 0.35s | 0.35s | 1.14s |
| 50k × 50 | 268.4s | 7.21s | 1.24s | 0.53s |
At 10,000 samples, FAISS CPU already delivers 30x speedup. The GPU backends show their strength at larger scales: at 50,000 samples, FAISS GPU achieves 216x speedup while cupy/cuML reaches 507x speedup. This crossover happens because GPU kernel launch overhead is amortized over more data, and cuML's brute-force search is highly optimized for batch queries.
Implementation
Building a fast, GPU-compatible KNN imputer required solving several technical challenges.
Automatic Backend Selection
FastKNNImputer automatically dispatches based on input array type.
Pass a numpy array, and it uses FAISS; pass a cupy array, and it uses cuML.
This is achieved using the array API, which provides a unified interface across array libraries.
This design ensures data stays on the GPU when using cupy - no expensive CPU transfers.
Why Not FAISS-GPU with cupy?
FAISS-GPU only accepts numpy arrays.
Even with faiss-gpu installed, passing cupy arrays would require transferring data to CPU, defeating the purpose.
cuML's NearestNeighbors operates natively on GPU memory, making it the right choice when your data pipeline is already GPU-resident.
Batched Operations
The naive approach processes rows one-by-one, which is catastrophic for GPU performance due to kernel launch overhead. Our implementation batches all operations:
# Batch prefill NaNs with fallback values
queries = xp.where(query_missing_mask, fallbacks, queries)
# Single search call for all rows
distances, indices = index.search(queries, self.n_neighbors)
# Vectorized neighbor retrieval: (n_rows, k, n_features)
all_neighbors = training_data[safe_indices]
# Vectorized weighted aggregation
weights = 1.0 / (distances + 1e-10)
imputed_rows = (all_neighbors * weights[:, :, None]).sum(axis=1) / weights.sum(axis=1, keepdims=True)
This eliminates thousands of Python loop iterations and GPU synchronization points.
Handling Sparse Missing Patterns
Real-world data rarely has uniform missingness.
When too few complete rows exist to build a reliable index, we iteratively exclude the most-NaN-heavy features until reaching a configurable threshold (min_data_ratio).
Features that cannot be imputed via KNN fall back to mean/median imputation with a warning.
Imputation Strategies
Three strategies are supported, matching scikit-learn's behavior:
- mean: Average of neighbor values
- median: Median of neighbor values
- weighted: Inverse-distance weighted average (closer neighbors contribute more)
Temporal Data Handling
For time series data in 3D format (samples × variables × timesteps), two modes are available:
- flatten: Treats all (variable, timestep) pairs as independent features. Fast, but allows future values to inform past imputations (temporal leakage).
- per_variable: Imputes each variable independently across its time dimension, preserving temporal causality at the cost of speed.
Installation & Usage
Install with optional GPU dependencies:
# CPU only (FAISS)
pip install "fknni[faisscpu]"
# With FAISS-GPU
pip install "fknni[faissgpu]"
# With RAPIDS (cuML, cupy)
pip install "fknni[rapids12]"
Usage mirrors scikit-learn:
from fknni import FastKNNImputer
import numpy as np
imputer = FastKNNImputer(n_neighbors=5, strategy="weighted")
# With numpy (uses FAISS)
X = np.random.rand(10000, 50)
X[np.random.rand(*X.shape) < 0.1] = np.nan
X_imputed = imputer.fit_transform(X)
# With cupy (uses cuML, stays on GPU)
import cupy as cp
X_gpu = cp.asarray(X)
X_imputed_gpu = imputer.fit_transform(X_gpu) # Returns cupy array
Conclusion
fknni brings modern hardware acceleration to KNN imputation.
For CPU-bound workloads, FAISS delivers 30-40x speedup out of the box.
For GPU pipelines or very large datasets, cuML pushes this to 500x.
The implementation handles real-world complexity—sparse missingness, configurable strategies, and automatic backend selection—while maintaining a scikit-learn compatible API.
The library is open source and available at github.com/Zethson/fknni.
If you're interested in engaging with me, consider following me on Bluesky or GitHub.
Acknowledgements
fknni was implemented by myself with major contributions from Nicolas Sidoux.
